Optimizing memory usage of Scikit-Learn models using succinct tries
We use the scikit-learn library for various machine-learning tasks at Zyte. For example, for text classification we'd typically build a statistical model using sklearn's Pipeline, FeatureUnion, some classifier (e.g. LinearSVC) + feature extraction and preprocessing classes. The model is usually trained on a developers machine, then serialized (using pickle/joblib) and uploaded to a server where the classification takes place.
Sometimes there can be too little available memory on the server for the classifier. One way to address this is to change the model: use simpler features, do feature selection, change the classifier to a less memory intensive one, use simpler preprocessing steps, etc. It usually means trading accuracy for better memory usage.
For text it is often CountVectorizer or TfidfVectorizer that consume most memory. For the last few months we have been using a trick to make them much more memory efficient in production (50x+) without changing anything from statistical point of view - this is what this article is about.
Let's start with the basics. Most machine learning algorithms expect fixed size numeric feature vectors, so text should be converted to this format. Scikit-learn provides CountVectorizer, TfidfVectorizer and HashingVectorizer for text feature extraction (see the scikit-learn docs for more info).
CountVectorizer.transform converts a collection of text documents into a matrix of token counts. The counts matrix has a column for each known token and a row for each document; the value is a number of occurrences of a token in a document.
To create the counts matrix CountVectorizer must know which column corresponds to which token. The CountVectorizer.fit method basically remembers all tokens from some collection of documents and stores them in a "vocabulary". Vocabulary is a Python dictionary: keys are tokens (or n-grams) and values are integer ids (column indices) ranging from 0 to len(vocabulary)-1.
Storing such a vocabulary in a standard Python dict is problematic; it can take a lot of memory even on relatively small data.
Let's try it! Let's use the "20 newsgroups" dataset available in scikit-learn. The "train" subset of this dataset has about 11k short documents (average document size is about 2KB, or 300 tokens; there are 130k unique tokens; average token length is 6.5).
Create and persist CountVectorizer:
from sklearn import datasets from sklearn.externals import joblib newsgroups_train = datasets.fetch_20newsgroups(subset='train') vec = CountVectorizer() vec.fit(newsgroups_train.data) joblib.dump(vec, 'vec_count.joblib')
Load and use it:
from sklearn.externals import joblib vec = joblib.load('vec_count.joblib') X = vec.transform(['the dog barks'])
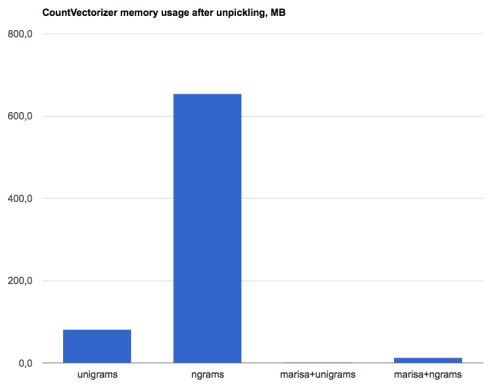
On my machine, the loaded vectorizer uses about 82MB of memory in this case. If we add bigrams (by using CountVectorizer(ngram_range=(1,2))) then it would take about 650MB - and this is for a corpus that is quite small.
There are only 130k unique tokens; it'll require less than 1MB to store these tokens in a plain text file ((6.5+1) * 130k). Maybe add an another megabyte to store column indices if they are not implicit (130k * 8). So the data itself should take only a couple of MBs. We may also have to somehow enumerate tokens and enable fast O(1) access to data, so there would be an overhead, but it shouldn't take 80+MB - we'd expect 5-10MB at most. The serialized version of our CountVectorizer takes about 6MB on disk without any compression, but it expands to 80+MB when loaded to memory.
Why does it happen? There are two main reasons:
- Python objects are created for numbers (column indices) and strings (tokens). Each Python object has a pointer to its type + a reference counter (=> +16 bytes overhead per object on 64bit systems); for strings there are extra fields: length, hash, pointer to the string data, flags, etc. (the string representation is different in Python < 3.3 and Python 3.3+).
- Python dict is a hash table and introduces overheads - you have to store hash table itself, pointers to keys and values, etc. There is a great talk on Python dict implementation by Brandon Rhodes, check it if you're interested in knowing more
Storing static string->id mapping in a hash table is not the most efficient way to do it: there are perfect hashes, tries, etc.; add Python objects overhead and here we are.
So I decided to try an alternative storage for vocabulary. MARISA-Trie (via Python wrapper) looked like a suitable data structure, as it:
- is a heavily optimized succinct trie-like data structure, so it compresses string data well
- provides a unique id for each key for free, and this id is in range from 0 to len(vocabulary)-1 - we don't have to store these indices ourselves
- only creates Python objects (strings, integers) on demand.
MARISA-Trie is not a general replacement for dict: you can't add a key after building, it requires more time and memory to build, lookups (via Python wrapper) are slower - about 10x slower than dict's, and it works best for "meaningful" string keys which have common parts (not for some random data).
I must admit I don't fully understand how MARISA-Tries work 🙂 The implementation is available in a folder named "grimoire", and the only information about the implementation I could find is Japanese slides which are outdated (as library author Susumu Yata says). It seems to be a succinct implementation of Patricia-Trie which can store references to other MARISA-Tries in addition to text data; this allows it to compress more than just prefixes (as in "standard" tries). "Succinct" means the Trie is encoded as a bit array.
You may never heard of this library, but if you have a recent Android phone it is likely MARISA-Trie is in your pocket - a copy of marisa-trie is in the Android 4.3+ source tree.
Ok, great, but we have to tell scikit-learn to use this data structure instead of a dict for vocabulary storage.
Scikit-learn allows passing a custom vocabulary (a dict-like object) to CountVectorizer. But this won't help us because MARISA-Trie is not exactly dict-like; it can't be built and modified like dict. CountVectorizer should build a vocabulary for us (using its tokenization and preprocessing features) and only then we may "freeze" it to a compact representation.
At first, we were doing it using a hack. fit and fit_transform methods were overridden: first, they call the parent method to build a vocabulary, then they freeze that vocabulary (i.e. build a MARISA-Trie from it) and trick CountVectorizer to think a fixed vocabulary was passed to the constructor, and then parents method is called once more. Calling fit/fit_transform twice is necessary because the indices learned on the first call and indices in the frozen vocabulary are different. This quick & dirty implementation is here, and this is what we're using in production.
I recently improved it and removed this "call fit/fit_transform twice" hack for CountVectorizer, but we haven't used this implementation yet. See https://gist.github.com/kmike/9750796.
The results? For the same dataset, MarisaCountVectorizer uses about 0.9MB for unigrams (instead of 82MB) and about 13.3MB for unigrams+bigrams (instead of 650MB+). This is a 50-90x reduction of memory usage. Tada!
The downside is that MarisaCountVectorizer.fit and MarisaCountVectorizer.fit_transform methods are 10-30% slower than CountVectorizer's (new version; old version was up to 2x+ slower).
Numbers:
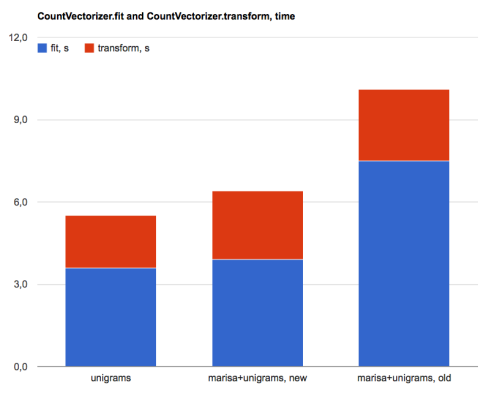
- CountVectorizer(): 3.6s fit, 5.3s dump, 1.9s transform
- MarisaCountVectorizer(), new version: 3.9s fit, 0s dump, 2.5s transform
- MarisaCountVectorizer(), old version: 7.5s fit, 0s dump, 2.6s transform
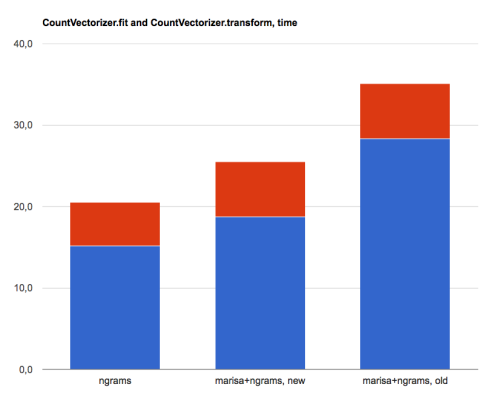
- CountVectorizer(ngram_range=(1,2)): 15.2s fit, 52.0s dump, 5.3s transform
- MarisaCountVectorizer(ngram_range=(1,2)), new version: 18.7s fit, 0.0s dump, 6,8s transform
- MarisaCountVectorizer(ngram_range=(1,2)), old version: 28.3s fit, 0.0s dump, 6.8s transform
'fit' method was executed on 'train' subset of '20 newsgroups' dataset; 'transform' method was executed on 'test' subset.
marisa-trie stores all data in a contignuous memory block so saving it to disk and loading it from disk is much faster than saving/loading a Python dict serialized using pickle.
Serialized file sizes (uncompressed):
- CountVectorizer(): 5.9MB
- MarisaCountVectorizer(): 371KB
- CountVectorizer(ngram_range=(1,2)): 59MB
- MarisaCountVectorizer(ngram_range=(1,2)): 3.8MB
TfidfVectorizer is implemented on top of CountVectorizer; it could also benefit from more efficient storage for vocabulary. I tried it, and for MarisaTfidfVectorizer the results are similar. It is possible to optimize DictVectorizer as well.
Note that MARISA-based vectorizers don't help with memory usage during training. They may help with memory usage when saving models to disk though - pickle allocates big chunks of memory when saving Python dicts.
So when memory usage is an issue, ditch scikit-learn standard vectorizers and use marisa-based variants? Not so fast: don't forget about HashingVectorizer. It has a number of benefits. Check the docs: HashingVectorizer doesn't need a vocabulary so it fits and serializes in no time and it is very memory efficient because it is stateless.
As always, there are some tradeoffs:
- HashingVectorizer.transform is irreversable (you can't check which tokens are active) so it is harder to inspect what a classifer has learned from text data.
- There could be collisions, and with improper n_features it could affect the prediction quality of a classifier.
- A related disadvantage is that the resulting feature vectors are larger than the feature vectors produced by other vectorizers unless we allow collisions. The HashingVectorizer.transform result is not useful by itself, it is usually passed to the next step (classifier or something like PCA), and a larger input dimension could mean that this subsequent step will take more memory and will be slower to save/load, so the memory savings of HashingVectorizer could be compensated by increased memory usage of subsequent steps.
- HashingVectorizer can't limit features based on document frequency (min_df and max_df options are not supported).
Of course, all vectorizers have their own advantages and disadvantages, and there are use cases for all of them. You can use e.g. CountVectorizer for development and switch to HashingVectorizer for production, avoiding some of HashingVectorizer downsides. Also, don't forget about feature selection and other similar techniques. Using succinct Trie-based vectorizers is not the only way to reduce memory usage, and often it is not the best way, but sometimes they are useful; being a drop-in replacement for CountVectorizer and TfidfVectorizer helps.
In our recent project, min_df > 1 was crucial for removing noisy features. Vocabulary wasn't the only thing that used memory; MarisaTfidfVectorizer instead of TfidfVectorizer (+ MarisaCountVectorizer instead of CountVectorizer) decreased the total classifier memory consumption by about 30%. It is not a brilliant 50x-80x, but it made the difference between "classifier fits into memory" and "classifier doesn't fit into memory".
Some links:
- MARISA-Trie based vectorizers - quick & dirty, use on your own risk;
- MARISA-Trie based CountVectorizer - not used in production, use on your own risk;
- fitting and transforming scripts used in benchmarks
- Google Spreadsheet with experiment results
There is a ticket to discuss efficient vocabulary storage with scikit-learn developers. Once the discussion settles our plan is to make a PR to scikit-learn to make using such vectorizers easier and/or release an open-source package with MarisaCountVectorizer & friends - stay tuned!